|
 |
||||||
|
 |
RESEARCH | ||||||
|
The Hay laboratory is firmly focused on establishing the role of ubiquitin and ubiquitin-like proteins in important biological processes. We presently have a number of exciting projects linking SUMO modification to ubiquitylation, in stress responses, DNA damage and arsenic therapy and in determining the mechanism of E3 ligase mediated conjugation.
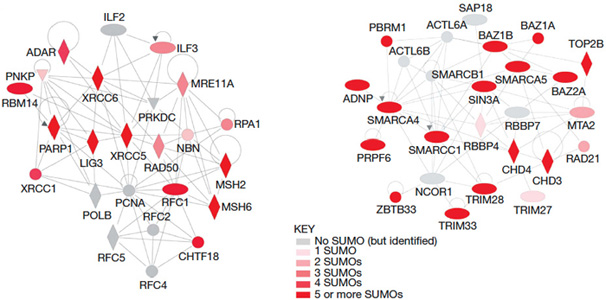
We are engaged in 4 related areas of research 1. Cellular regulation by SUMO DNA-damage signaling utilizes a variety of posttranslational modifiers as molecular switches to regulate the signaling network. Ubiquitin and, more recently, Small Ubiquitin-Like Modifier (SUMO) have been shown to be important mediators of this response. We demonstrated (Bruderer et al., 2011) that RNF4, a highly conserved small ubiquitin-like modifier (SUMO)-targeted ubiquitin E3 ligase could interact with extensive networks of SUMO modified proteins involved in chromatin remodeling and DNA repair (Fig. 1), RNF4 is a novel DNA damage-responsive protein that plays a role in homologous recombination and integrates SUMO modification and ubiquitin signaling in the cellular response to genotoxic stress (Yin et al., 2012; Maure et al., 2016).
The small ubiquitin-like modifier (SUMO), initially characterized as a suppressor of a mutation in the gene encoding the centromeric protein MIF2, is involved in many aspects of cell cycle regulation. We have used Caenorhabditis elegans to establish the contribution of SUMO to a timely and accurate cell division. (Pelisch et al, 2014). During Caenorhabditis elegans oocyte meiosis, a multi-protein ring complex (RC) localized between homologous chromosomes, promotes chromosome congression. We demonstrated that dynamic SUMO modification and the presence of SIMs in RC components (Fig. 2) generate a SUMO-SIM network that facilitates assembly of the RC (Pelisch et al, 2017). 

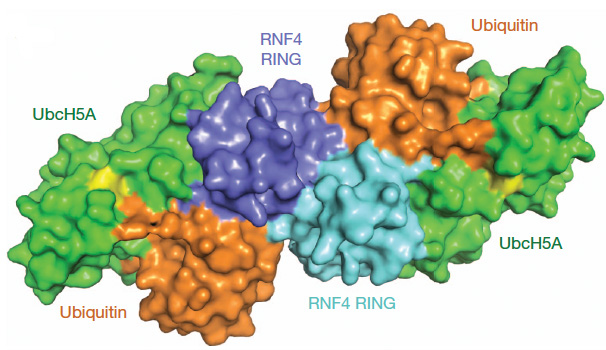
3. Determining the mechanism of RING mediated ubiquitin modification. Ubiquitin modification is achieved by the sequential action of 3 enzymes: an E1 activating enzyme that links ubiquitin to a cysteine residue in an E2 conjugating enzyme and E3 ubiquitin ligases that catalyse the transfer of the ubiquitin from the E2~ubiquitin onto the substrate protein. There are more than 600 human genes that encode ubiquitin E3 ligases and as they influence almost all aspects of biological activity they often play critical roles in the development of disease. By far the most common E3 ligases belong to the RING family but how they stimulate ubiquitin transfer was a long-standing mystery. This mystery was solved by our determination of the crystal structure of the RNF4 RING E3 ligase bound to ubiquitin linked E2. This gives a view of an E3 ligase, E2~ubiquitin complex primed for catalysis (Fig. 4) and suggests a unified mechanism for ubiquitin transfer that could apply to most other E3 enzymes (Plechanovova et al, 2011; Plechanovová et al, 2012).
Fig. 3 Structure of the RNF4 RING bound to ubiquitin-loaded UbcH5A. Surface representation of the complex. Individual RING protomers are coloured cyan and blue, UbcH5A is green, ubiquitin is orange and the isopeptide linkage between the C terminus of ubiquitin and K85 of UbcH5A is shown in yellow. To establish how the SIMs in RNF4 recognised SUMO chains we collaborated with the NMR group of Steve Matthews and obtained a structural model that indicated how multiple SUMO substrates were recognised by the E3 ubiquitin ligase (Xu, Plechanovová et al 2014). It was also established how substrates are recognized in the context of ubiquitin chain sysnthesis (Branigan et al., 2015). We demonstrated that SUMO chains were not only substrates for RNF4 but were activators of ubiquitin ligase activity. In the absence of chains RNF4 is monomeric and inactive, but chains induce dimerization and thus ubiquitin ligase activity (Rojas-Fernandez et al, 2014).
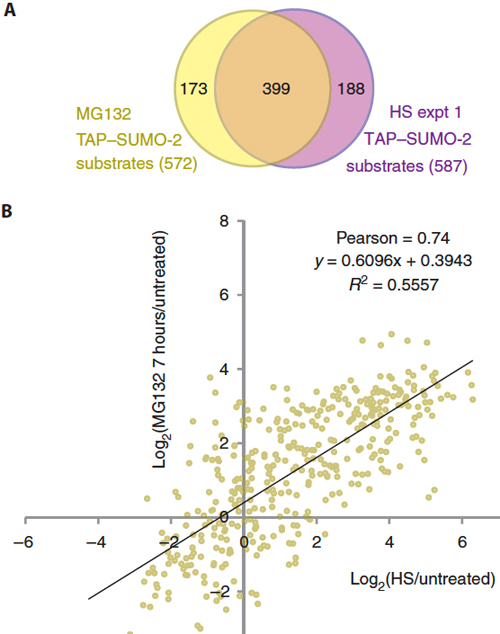
4. System wide analysis of SUMO modification in response to stress To establish the relationship between SUMO and protein degradation by the proteasome, we performed a quantitative proteomic analysis of SUMO-2 substrates after short- and long-term inhibition of the proteasome with MG132. Comparisons with changes to the SUMO-2 conjugate subproteome in response to heat stress (Golebiowski et al, 2009; Bruderer et al, 2011) revealed qualitative and quantitative parallels between both conditions (Fig. 5) However, in contrast to heat stress, the MG132-triggered increase in SUMO-2 conjugation depended strictly on protein synthesis, implying that the accumulation of newly synthesized, misfolded proteins destined for degradation by the proteasome triggered the SUMO conjugation response (Tatham et al, 2011). These findings, together with ChIpSeq analysis to define the chromatin landscape of SUMO modification (Seifert et al, 2015) suggest multiple, proteasome-independent roles for SUMOs in the cellular response to the accumulation of misfolded proteins. Although proteomic studies have identified hundreds of sumoylated substrates, methods to identify the modified lysines on a proteomic scale were lacking. We developed a method that enabled proteome-wide identification of sumoylated lysine residues and we can now identify thousands of sites of SUMO modification in a single experiment. This technology can be adapted for use with any Ubl and has revolutionized Ubl proteomics (Tammsalu et al, 2014).
References
Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. (2011) Purification and identification of endogenous polySUMO conjugates. EMBO Reports. 12:142-8.
Geoffroy MC, Jaffray EG, Walker KJ, Hay RT. (2010) Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Molecular Biology of the Cell 21: 4227-39.
Pelisch F, Sonneville R, Pourkarimi E, Agostinho A, Blow JJ, Gartner A and Hay RT (2014) Dynamic SUMO modification regulates mitotic chromosome assembly and cell cycle progression in Caenorhabditis elegans. Nature Communications 5: 5485. Pelisch F, Tammsalu T, Wang B, Jaffray EG, Gartner A, Hay RT (2017) A SUMO-Dependent Protein Network Regulates Chromosome Congression During Oocyte Meiosis. Molecular Cell 65:66-77 Plechanovová A, Jaffray E, Tatham MH, Naismith JH, Hay RT. (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115-120
Seifert A, Schofield P, Barton GJ and Hay RT (2015) Proteotoxic stress reprogrammes the chromatin landscape of SUMO modification. Science Signaling 8: rs7
Tammsalu T, Matic I, Jaffray EG, Ibrahim AFM, Tatham MH and Hay RT (2014) Proteome-wide Identification of SUMO2 Modification Sites. Science Signaling 7: rs2. |
|||||||
Updated 15/08/17 |
back to top |