News
The results from Giulia's biomolecular simulations explain experimental observations made by our long-standing collaborators Arnaud Javelle and Anna-Maria Marini to yield key insights into the variations of substrate specificity and the type of mechanism for transporting ammonium in Mep-Amt-Rh proteins. Importantly, only one of these, Mep2, is capable of triggering filamentation in yeast, leading to infection. The findings have been published in mBio.
We are happy to announce that two new BBSRC-Eastbio funded PhD students have just joined the group, Alp Tegin Şahin and Tim Spankie - welcome to Scotland!
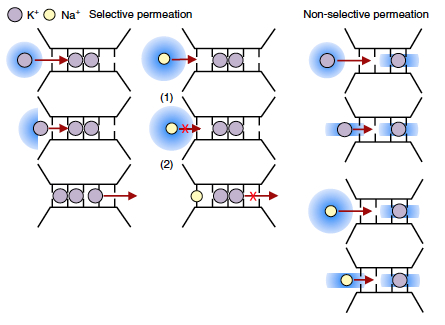
Together with Wojciech, Bert and Andrei we summarise the current consensus - and disagreements - on the permeation mechanism at play in K+ channels underpinning their high permeation efficiency and strict ion selectivity in J. Mol. Biol. We review recent and older experimental and computational findings that may contribute to solving this problem, eventually!
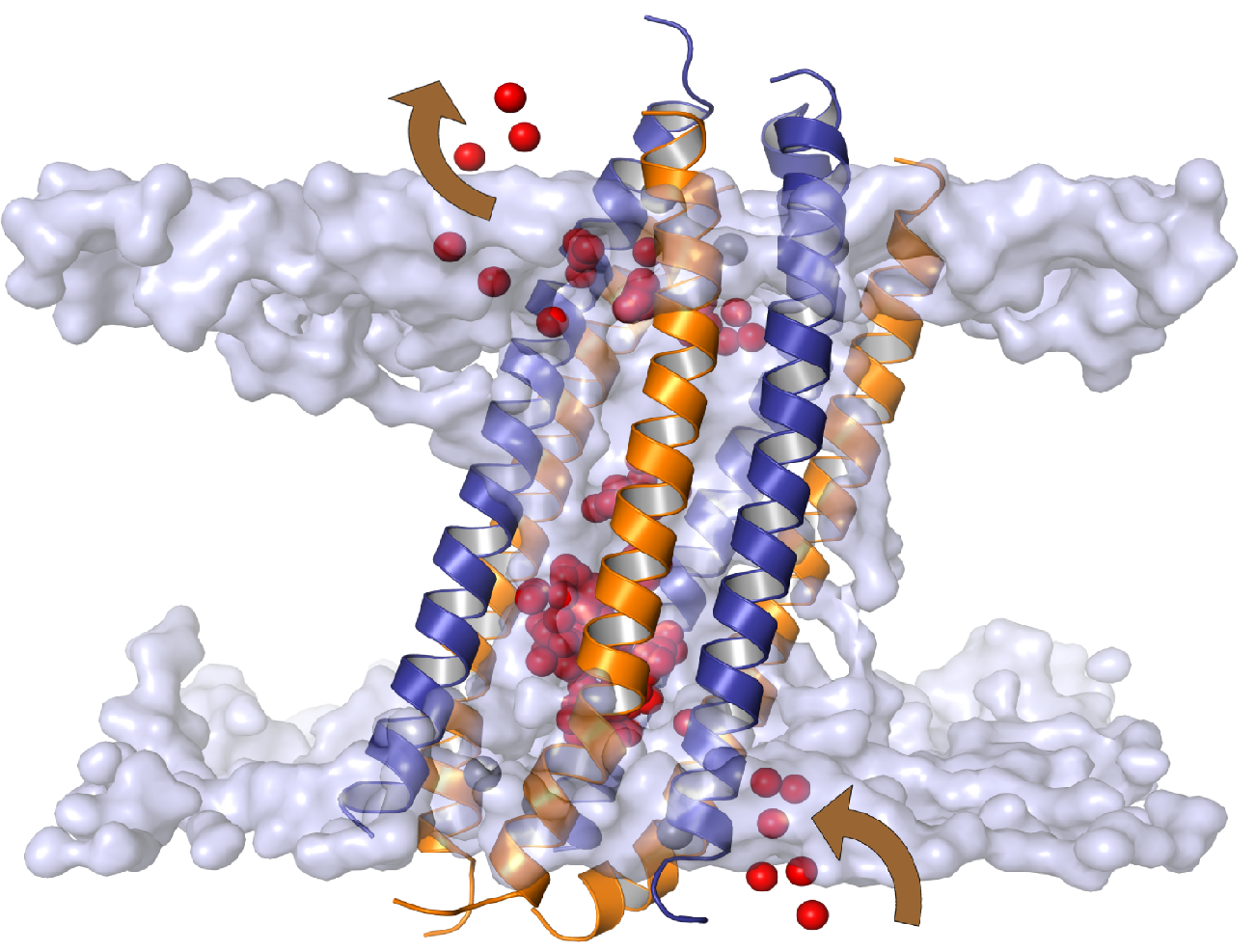
Giulia's and Marcus' simulations, together with experimental results from our collaborators Arnaud Javelle and Paul Hoskisson (University of Strathclyde) and Anna-Maria Marini (Free University of Brussels), have revealed a novel "two-lane" mechanism of ammonium transport in bacterial and eukaryotic ammonium transporter, now published in eLife. Also see the insightful commentary article by William Allen & Ian Collinson on the significance of the findings.
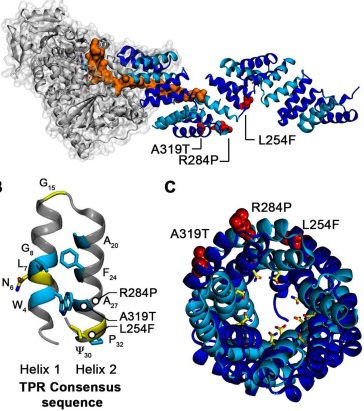
Salomé's work on mutations in the TPR domain of OGlcNac Transferase that are linked to
Intellectual Disease shows that they change the dynamics and elasticity of the domain,
with important implications for its ability to bind substrates. You can read the full
story open access here:
We are welcoming Dominik Gurvič and Callum Ives, who will join the group in early September
and work alongside BBSRC-Eastbio PhD student Neil Thomson on MRC-funded PhD projects in the fields
of ion channels and antibacterial resistance.
Our collaborative work with Claudia Steinem's group from Göttingen shows how the beta-lactam ampicillin binds to PorB from
N. meningitidis or meningococca, the causative agent of bacterial meningitis, with many important implications for antibiotic design.
The study is now out in Scientific Reports.
Our work showing why potassium channels are so selective for the conduction of only K+ ions has just been published in
Nature Chemistry. See also the
News & Views article by Ben Corry in the same issue.
Research Focus
Molecular Simulations of Membrane Protein Function
Biological cells enable the processes of life to occur within small, highly controlled compartments far from equilibrium. Cells are bounded
by biomembranes, which are impermeable to many solutes and present a barrier to the exchange of matter and information between the outside and
the inside of the cell. Since life requires a constant flux of matter and energy, specialized proteins have evolved
that enable the transfer of ions, molecules, and signals between the cell and the external world. These processes are of such central importance for
the life of cells that, in humans, about one-third of the genome encodes membrane proteins and almost one-half of all marketed drugs target membrane proteins.
A core interest of the group are the molecular mechanisms of membrane protein function, their interaction with drugs and substrates, and their wider
environment inside lipid bilayers. Key present examples are membrane ion channels (e.g. the ion conduction efficiency and selectivity of potassium channels), pores
in the outer membrane of bacteria that are found to be mutated in bacterial strains resistant to antibiotics (e.g., neisserial PorB), membrane surface receptors such as G-protein coupled receptors (e.g. muscarinic and opioid receptors), and membrane transporters such as multidrug efflux pumps or AmtB. Often, we are especially interested in the role of realistic membrane potentials in the function of these proteins.
Biomechanics of Tandem Repeat Proteins
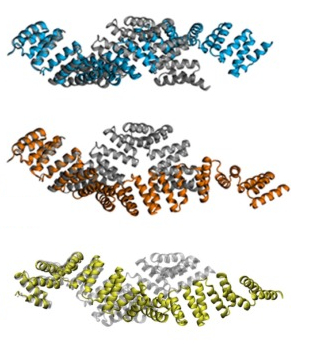
Proteins form the machinery of biological cells. A large proportion of the proteome in higher organisms consists of
solenoid repeat proteins, in which small conserved structural units stack to yield extended structures with large
water-exposed surfaces. These proteins often play a role in enabling tight protein-protein binding interactions
or they can serve as structural scaffolds.
Depending on the fundamental repeating unit, they can be classified as HEAT, leucine-rich, tetratricopeptide, ankyrin or armadillo repeat proteins.
In alpha-helical solenoid proteins, the building blocks usually consist of two to three alpha-helices.
Alpha-solenoids such as HEAT repeat proteins are often exceptionally
flexible and elastic, features that
are key to their biological functions.
In recent years, we have focused on tetratricopeptide (TPR) domains (shown here), studying their
biomechanics and the
role of changes in TPR dynamics in human disease.
Join the Team
Several PhD studentships in the area of computational biophysics, molecular modelling and simulations as well as chemoinformatics/machine learning
are available in the group.
The positions are fully funded through MRC, BBSRC and Wellcome Trust PhD programmes.
For more information, see Jobs and follow the link there.
We are looking for biochemists, chemists, biologists and physicists interested in working at the interface between the traditional
disciplines. Contact us if you are interested in studying the mechanisms that drive biology with a view to developing drugs - and if you like computational work.
For a clearer picture, read more about our past and present research and have a look at our publications.
Further Information:
UZ Group Twitter profile.
Google Scholar profile (PI).
Contact Information.